Recombinant Expression Validates De Novo Sequencing Accuracy: Trastuzumab & Anti-Factor XII Case Study
Husseini, I.3, Ruiz, A.3, Mitchell, M.1, Wenzel, R.2, Nazabal, A.3, Feldman, H.M.1
1Green Mountain Antibodies, Burlington, VT 05401
2 CovalX Analytics Inc., Woburn, MA 01801
3 CovalX Analytics SA, Martillac, France 33650
Introduction
In this case study, we evaluated the accuracy and functionality of sequences derived from two reference antibodies: anti-HER2 (trastuzumab) and anti–Factor XII (GMA-141) through recombinant expression and de novo sequencing. De novo sequencing via mass spectrometry enables recovery of protein sequences directly from purified protein material, bypassing the need for genetic source material. Optimization of transient transfection improved expression levels, resulting in yield and quality suitable for scale-up. Functional assays, including ELISA and biolayer interferometry, confirmed target binding consistent with reference antibodies. These results support de novo sequencing as a reliable approach for generating functional recombinant antibodies directly from purified monoclonals for research and therapeutic applications.
Background
Monoclonal antibodies are indispensable tools in biomedical research and therapeutic development. Traditional sequencing methods rely on access to hybridoma cell lines, B cells, or mRNA. De novo protein sequencing, using mass spectrometry, enables sequence recovery directly from purified antibodies. However, functional verification of these sequences is essential. This case study evaluates the accuracy of de novo-derived sequences through recombinant expression and characterization of two well-established antibodies.
Objectives
- Assess the accuracy of de novo sequencing in recovering functional antibody sequences.
- Optimize expression and evaluate yield and quality for recombinant scale-up feasibility.
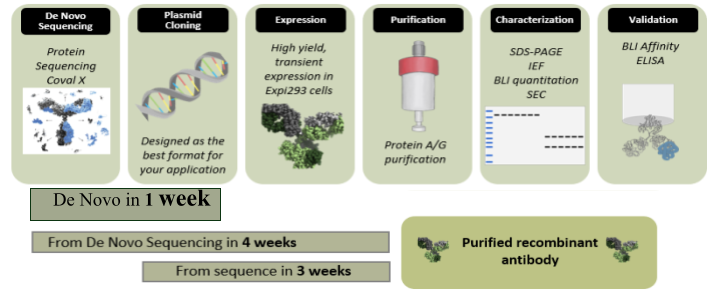
Process Overview
Methods
- Reference Antibodies & De Novo Sequencing
Reference trastuzumab for de novo sequencing was obtained from the National Institute of Standards and Technology (NIST). Reference GMA-141 was obtained as a purified antibody produced from a monoclonal hybridoma cell line from Green Mountain Antibodies. A high-mass MALDI MS analysis (Bruker Autoflex II with CovalX HM5 interaction module) was performed to verify reference integrity (Fig 1A). For de novo sequencing, the reference samples were submitted to Trypsin, Chymotrypsin, Asp-N, Elastase, Lys-C, Glu-C and αLP proteolysis followed by liquid chromatography (Ultimate 3000-RSLC) and nLC-Orbitrap Fusion Lumos MS/MS analysis. The resulting peptide sequences were designed into an overlap map with 100% sequence coverage (Fig 1B).
- Construct Design & Expression
Heavy and light chain sequences were codon optimized for expression in expi293 cells with a leader sequence added for secretion. Genes were synthesized and cloned into mammalian expression vectors. The heavy and light chain plasmids were transiently transfected at three different ratios (1:1, 1:2, and 1:4) to identify the optimal expression ratio. Supernatant was harvested at seven days and yield quantified using Biolayer Interferometry (BLI) on a GatorPlus. Supernatant was bound to anti-hFc or anti-mFc probes and concentration extrapolated from a standard curve of human or mouse IgG. Supernatant from all expression ratios were pooled for purification on a Protein G (GMA-141) or Protein A (trastuzumab) column. Samples were eluted in 0.1M glycine, pH 2.8, and buffered with tris, pH 7.4 then dialyzed to PBS (pH 7.4) and filtered with a 0.22 um filter. Protein yield was quantified using absorbance at 280 nm with an extinction coefficient of 1.4.
- Antibody Characterization
Purified recombinant antibodies were analyzed by SDS-PAGE (4-12% Bis/Tris gel) and size exclusion chromatography (AKTA Pure 25L). For indirect ELISA, purified recombinant or reference antibody was titrated against antigen-coated ELISA plates (HER2 [ACRO, HE2-H5253] or Factor XII). Bound antibody was detected using goat-anti-mIgG-Fcy-HRP or goat-anti-huIg-Kappa-HRP with OPD substrate. Apparent affinity constant (KD) for binding to Factor XII or HER2 was measured using BLI on a GatorPlus. Antibodies were bound to probes and open binding sites blocked with Fc fragments (anti-HER2 only). Probes were washed and placed into serial dilutions of Factor XII or HER2 and the association and dissociation were measured. Affinity was determined by the 1:1 local fitting software algorithm using derived kinetic parameters for each analyte concentration.
Results
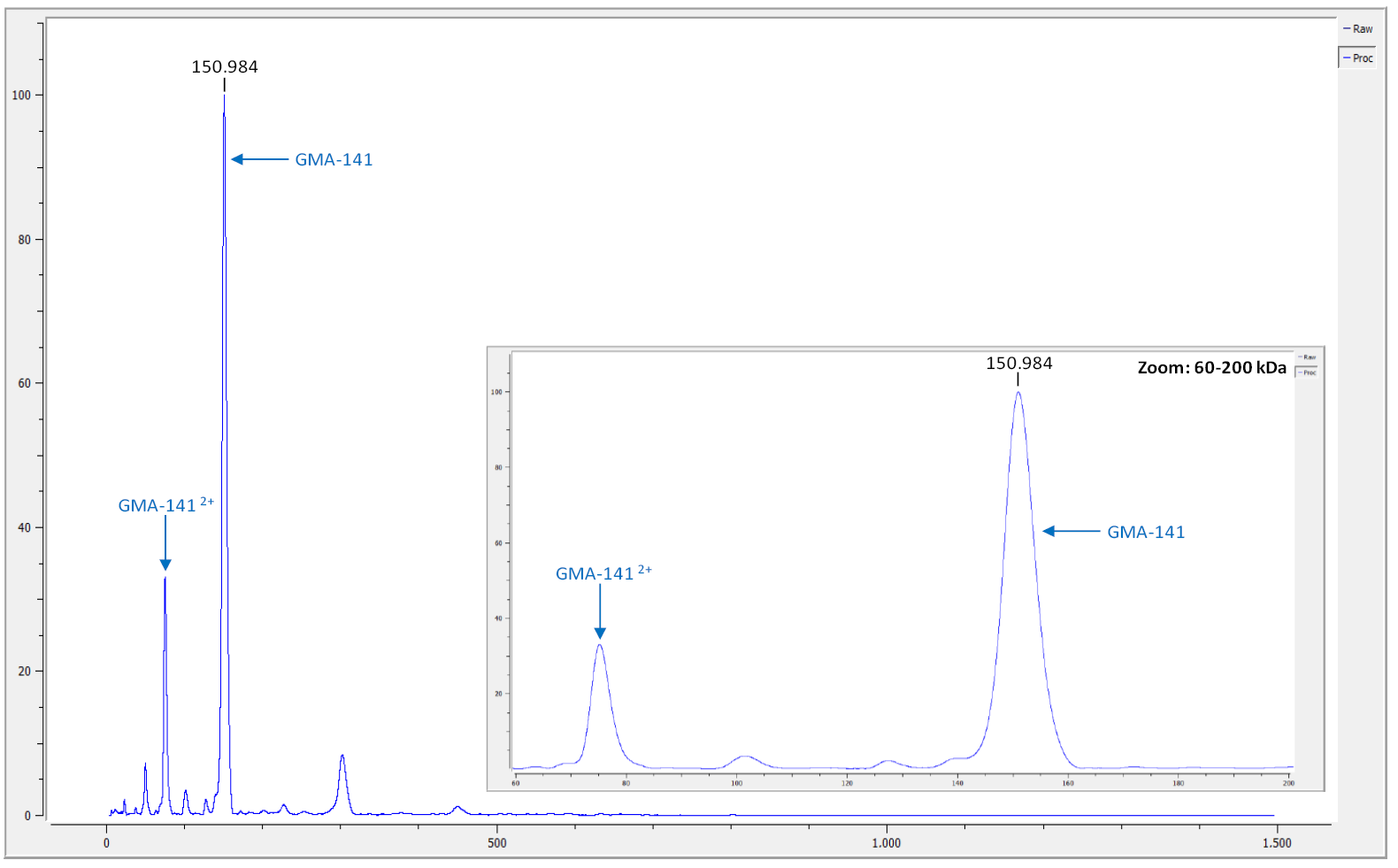
Before beginning De Novo sequencing analysis the intact protein sample is first interrogated by high mass MALDI analysis to confirm protein integrity, detect impurities and any significant fragmentation (Fig 1A). After high mass MALDI confirmation, the protein is digested with 7 separate enzymes allowing thorough peptide coverage (Fig 1B – legend). Each of the seven separate peptide mixtures is then analyzed by nano-liquid chromatography electrospray mass spec detection with MS/MS fragmentation and peptide identification. The latest EThCD dissociation is utilized to allow direct fragmentation detection of all amino acid residues including direct discernment of Isoleucine/Leucine residues. Several PTM’s are reported with N-glycan detection and percentages included (not shown here).
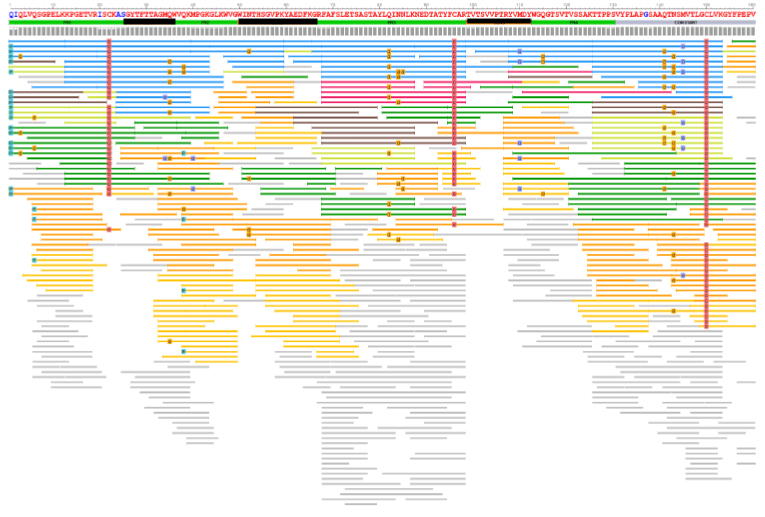
Figure 1: Expression optimization and biophysical characterization of de novo–derived antibodies.
(A) Intact analysis by High Mass MALDI for initial QC of antibody sample. (B) [Partial Heavy Chain shown] Sequence coverage of heavy chain from GMA-141 antibody sequenced. CDR framework identified, as well as each peptide from 7 different enzymes presented. Several PTMs are defined and N-glycan coverage detected.
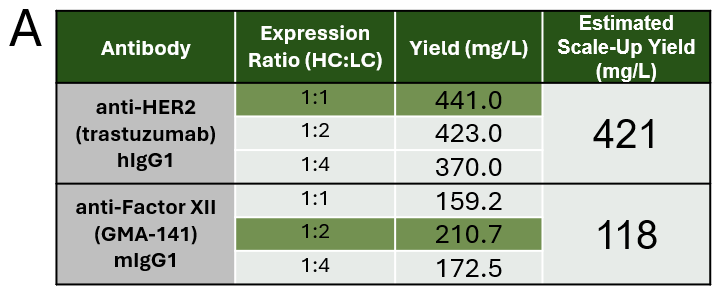
Figure 2: Expression optimization and biophysical characterization of de novo–derived antibodies.
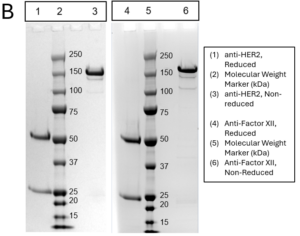
- A) Quantitation of antibody expression by biolayer interferometry (BLI) across different heavy-to-light chain transfection ratios. Optimal ratio for each antibody highlight in green. Estimated scale-up yield is extrapolated from purification yield assuming the optimized transfection parameters. (B) SDS-PAGE analysis of purified recombinant antibody under reducing and non-reducing conditions. (C) Size-exclusion chromatography (SEC) profiles used to evaluate monomer content and detect potential aggregation. (D) Isoelectric focusing (IEF) analysis to determine the isoelectric point (pI) of the purified antibodies.
Results Continued
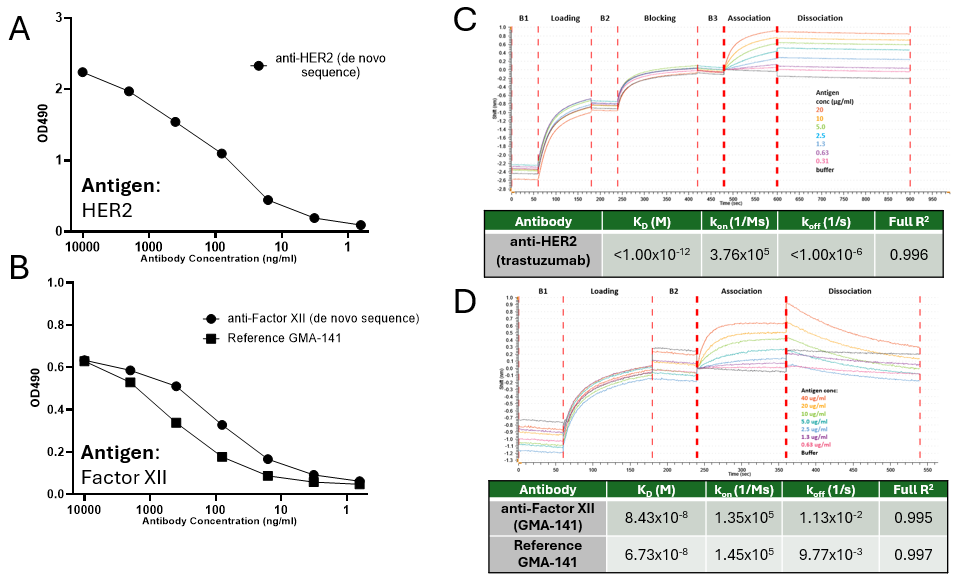
After the full amino acid sequences have been determined, rapid expression of antibodies can be conducted, by first optimizing heavy and light chain ratios for optimal expression (1:1 for anti-HER 2 and 1:2 ratio for anti-Factor XII – Fig 2A). This process generated 12.4mg of anti-HER2 and 3.1mg of anti-Factor XII for an estimated scale-up of 421mg/L and 118mg/L, respectively. After purification, antibodies are characterized by SDS-PAGE, SEC and IEF (Fig 2B, C &D) to confirm protein presence, aggregation levels and pI values. Finally, binding of each antibody is confirmed through functional assays of ELISA and BLI (Fig 3A-D) generating recombinant trastuzumab which exhibited a KD of <1.00 pM (*When koff is <1.00 x10-6 s ⁻¹, KD can only be approximated), consistent with reported high-affinity binding. Recombinant GMA-141 showed a KD of 84.3 nM, closely matching the reference antibody (KD = 67.3 nM), supporting functional equivalence.
Figure 3: Functional characterization of de novo–derived antibodies by ELISA and biolayer interferometry.(A,B) ELISA binding curves for recombinant trastuzumab (A) and GMA-141 (B), demonstrating specific binding to the target antigen.(C,D) BLI sensorgrams and kinetic analyses for recombinant trastuzumab (C) and GMA-141 (D).
Conclusions
- De novo sequencing can rapidly and accurately recover functional antibody sequences directly from purified protein materials such as antibodies.
- Recombinant expression provides a robust platform for the rapid validation of sequence information and generation of viable new antibody material.
- This workflow supports rapid recovery of legacy antibodies and generation of research or therapeutic reagents without relying on original cell lines.
Acknowledgments
We gratefully acknowledge the collaborative efforts of both Green Mountain Antibodies and CovalX in the successful execution of this project. We extend special thanks to the technical teams at both organizations, whose expertise and dedication were instrumental in generating high-quality data and maintaining project momentum.
For questions or inquiries:
- Green Mountain Antibodies: Carrie Rice, Director of Business Development at Sales@greenmoab.com or visit www.greenmoab.com
- CovalX: Ryan Wenzel, PhD; Director of N.A. Operations at ryan.wenzel@covalx.com or visit www.covalx.com